Proteins
Quick Notes
- Proteins are polymers made up of amino acid monomers joined by peptide bonds (−CONH−).
- Proteins have complex shapes that are described in terms of a Primary, Secondary and Tertiary structure.
- Primary structure refers to the sequence of amino acids bonded together.
- Secondary structure includes α-helices and β-pleated sheets, stabilised by hydrogen bonding between C=O and N−H groups from different amino acids.
- Tertiary structure is the 3D folding of proteins, stabilised by hydrogen bonds, disulfide bonds (S−S), ionic bonds, and hydrophobic interactions between R groups on different amino acids.
- Hydrolysis of proteins breaks peptide bonds, forming the constituent amino acids.
- Amino acids can be separated and identified by thin-layer chromatography (TLC).
- As amino acids are colourless, ninhydrin or UV light is used with a chromatogram, and amino acids are identified by Rf values.
Full Notes
Structure of Proteins
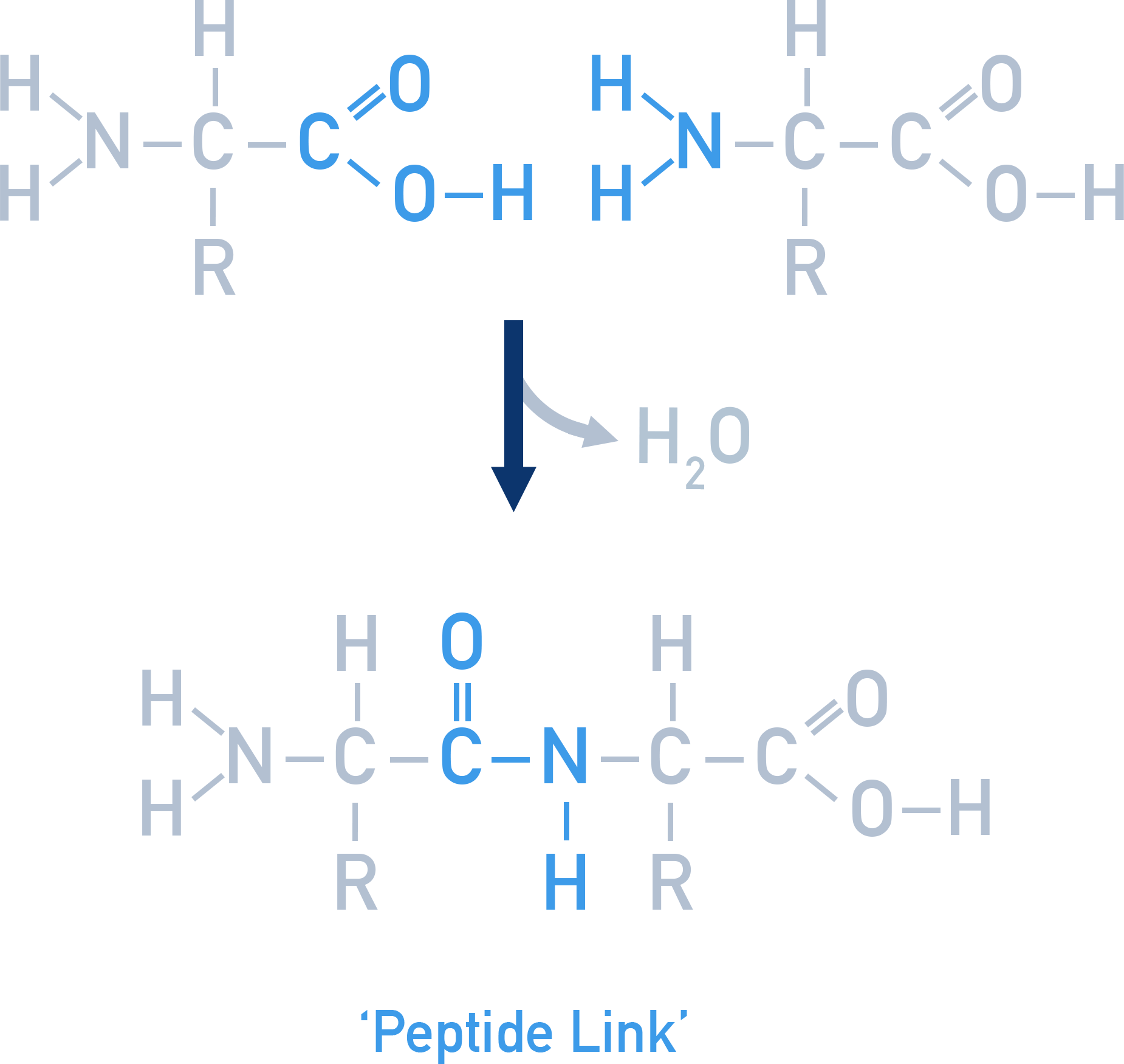
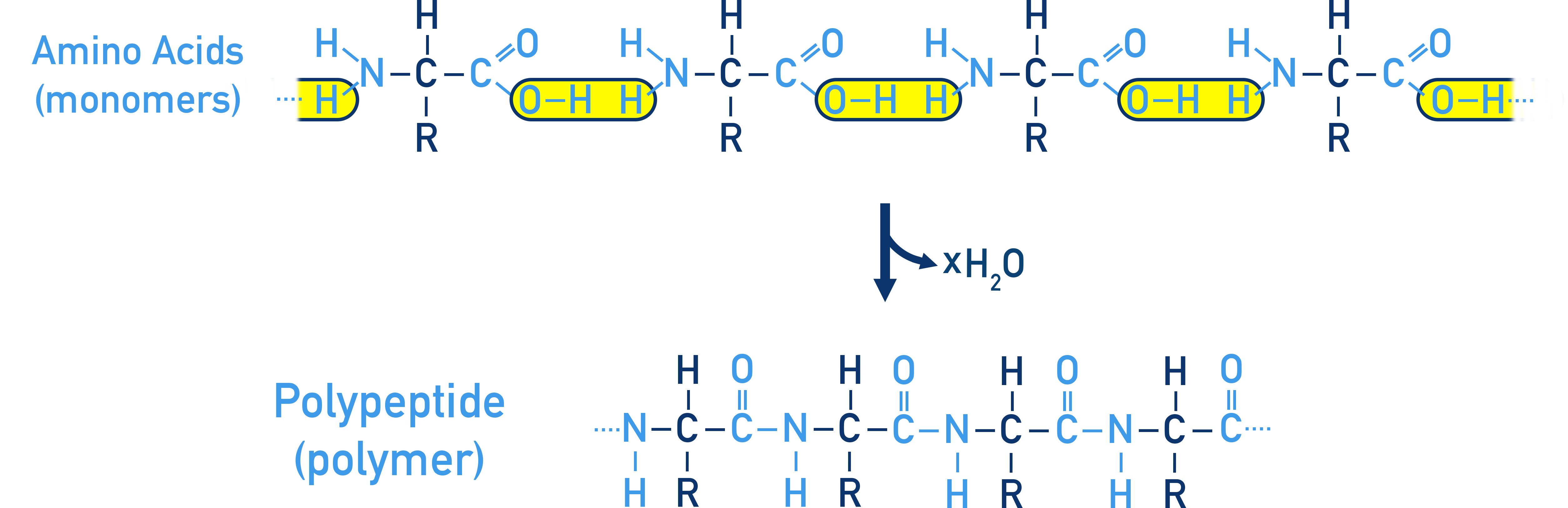
Proteins are naturally occurring polymers of amino acids linked by peptide bonds (−CONH−).
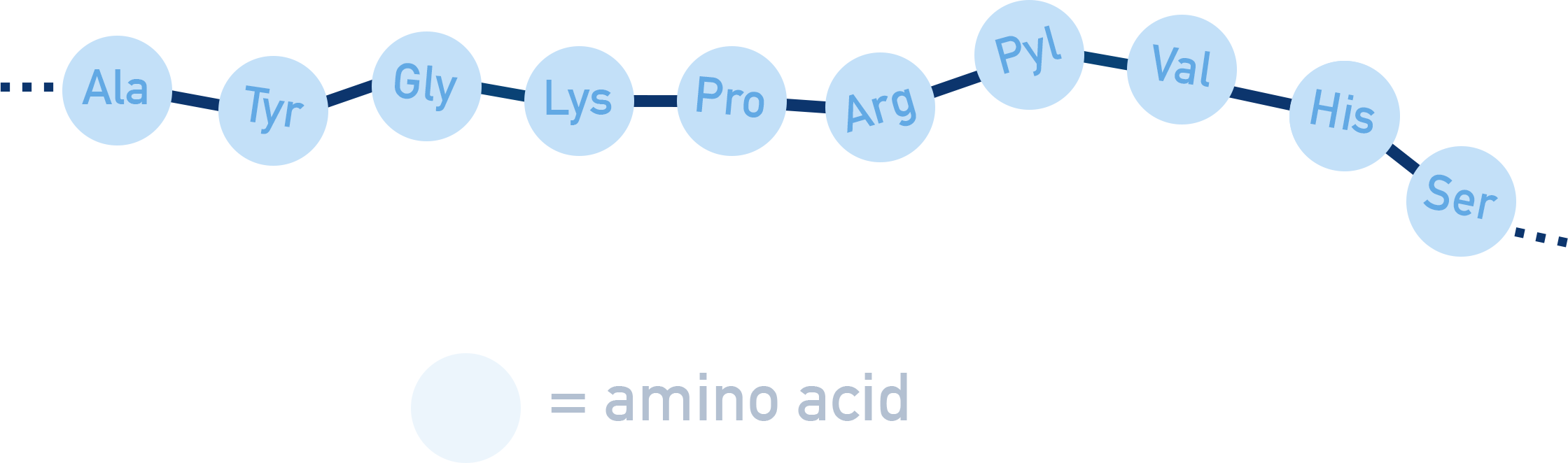
Peptide bonds form by condensation reactions between amino acids, releasing water:
The process can keep happening between more amino acids, forming a polypeptide chain (polymer).
Peptide bonds can be broken apart in hydrolysis reactions. The hydrolysis of proteins (with acid or alkali) releases the amino acids that made up the protein .
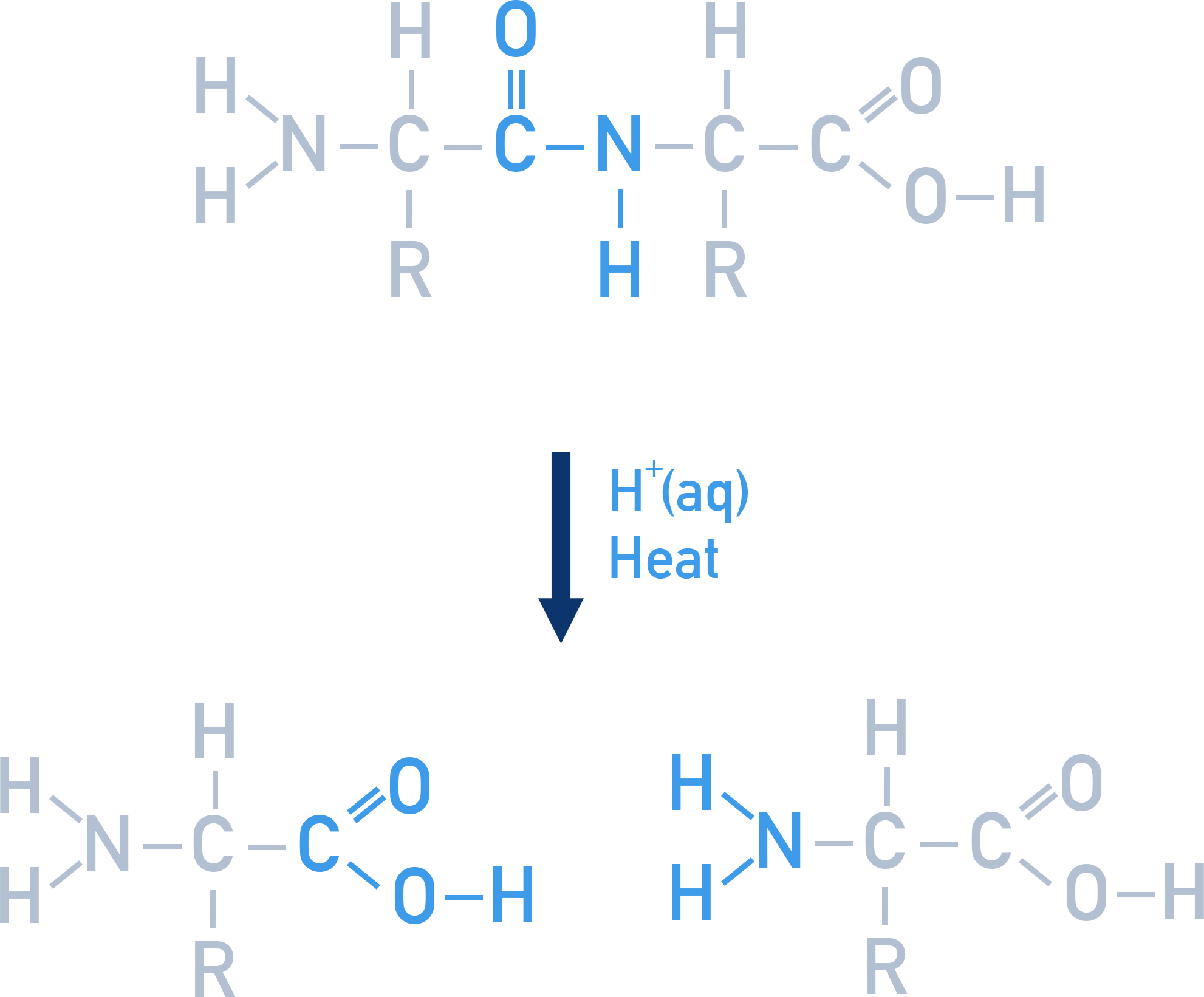

The conditions used for hydrolysis determine the form of the amino acids released.
- If acidic conditions are used, then the amino acids may exist as positively charged ions with the NH2 group accepting a H+ ion to form NH3+
- If alkaline conditions are used, then the carboxylic acid groups may be forced to lose H+ ions and exist as carboxylate ions (COO−)
Levels of Protein Structure
Proteins can have very complicated shapes that are difficult to analyse. We describe the structure of proteins in terms of a Primary, Secondary and Tertiary structure. (Proteins made up of more than one polymer chain also have a quaternary structure, however you don’t need to learn that for AQA A-Level Chemistry!).
Primary Structure
The primary structure is the sequence of amino acids in a polypeptide chain.
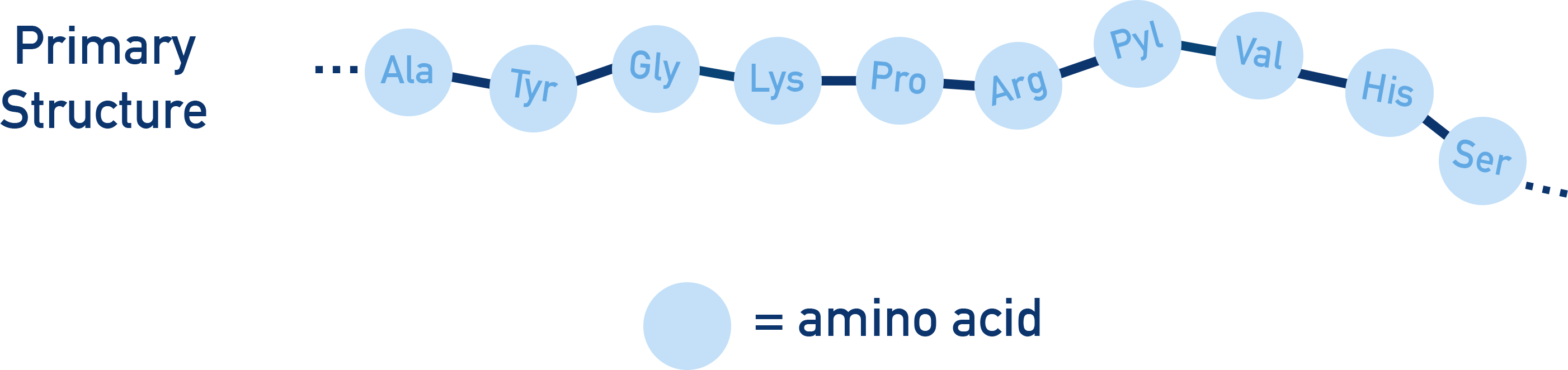
Determines the overall structure and function of the protein.
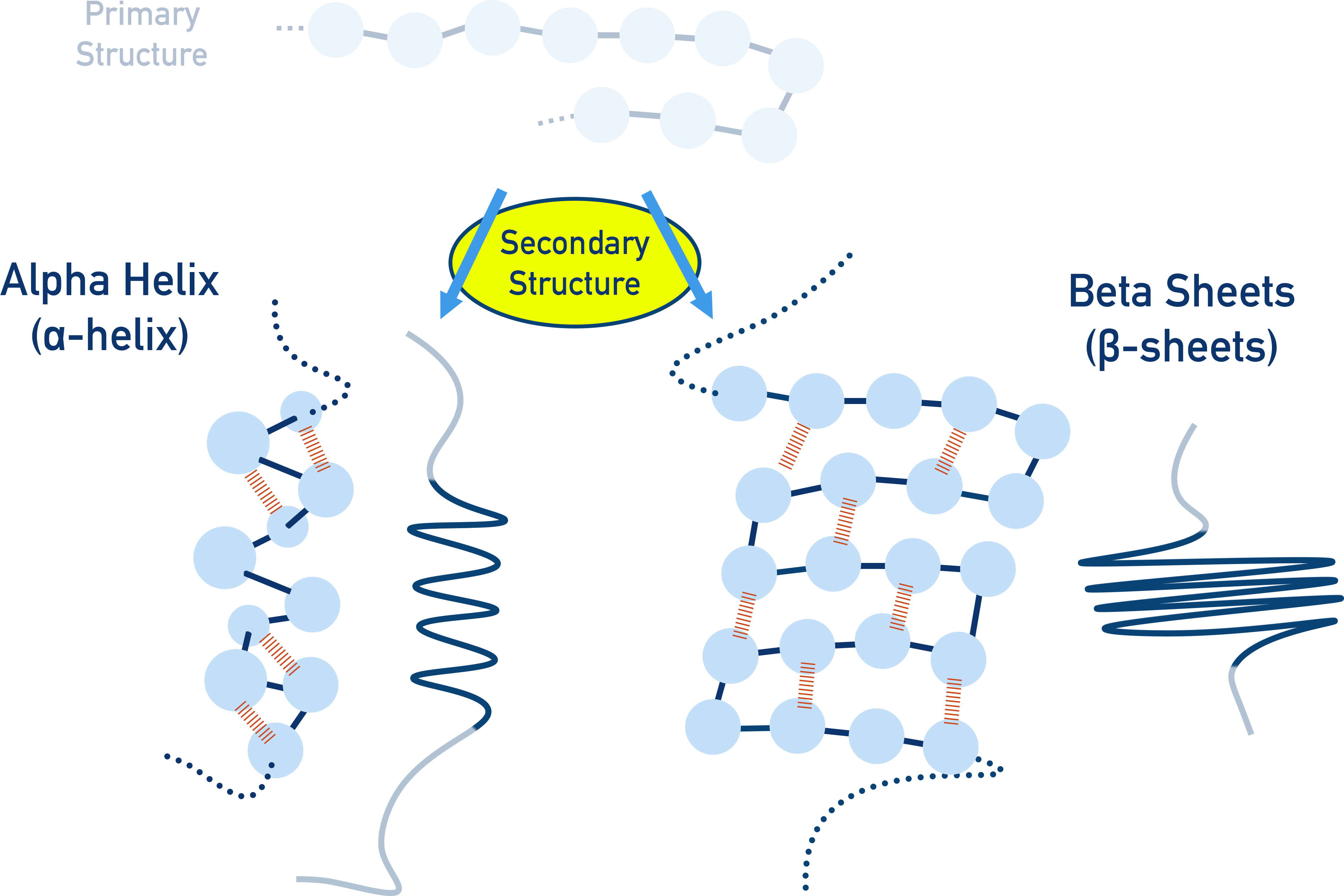
Secondary Structure
The secondary structure describes two types of shape (α-helix and β-pleated sheet) that arise from hydrogen bonding between C=O and N−H bonds from different amino acids in the polymer chain
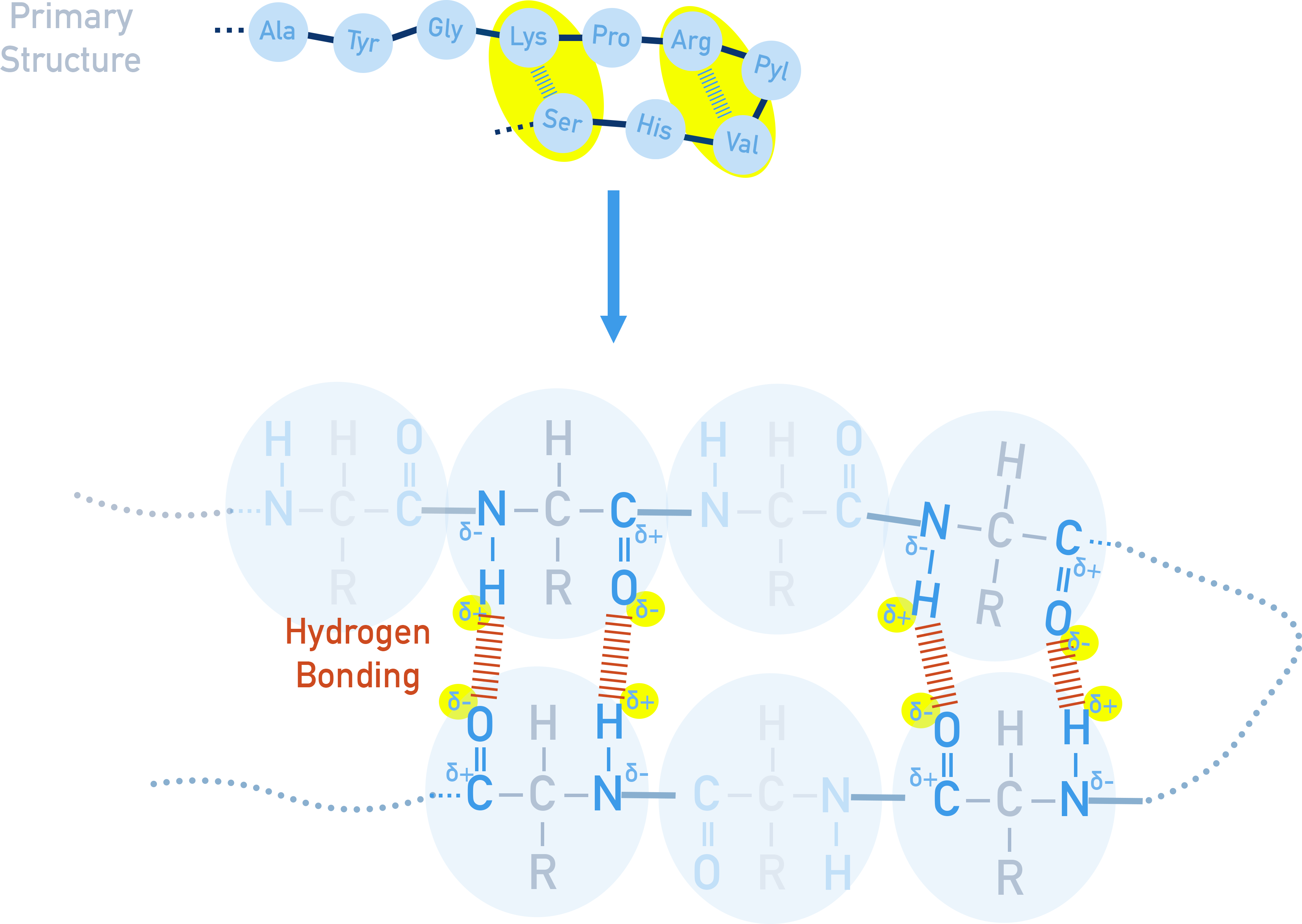
α-helix – a spiral structure stabilised by hydrogen bonds
β-pleated sheet – strands linked side by side with hydrogen bonds
Tertiary Structure
The tertiary structure describes the 3D folding of the protein due to interactions between R groups.
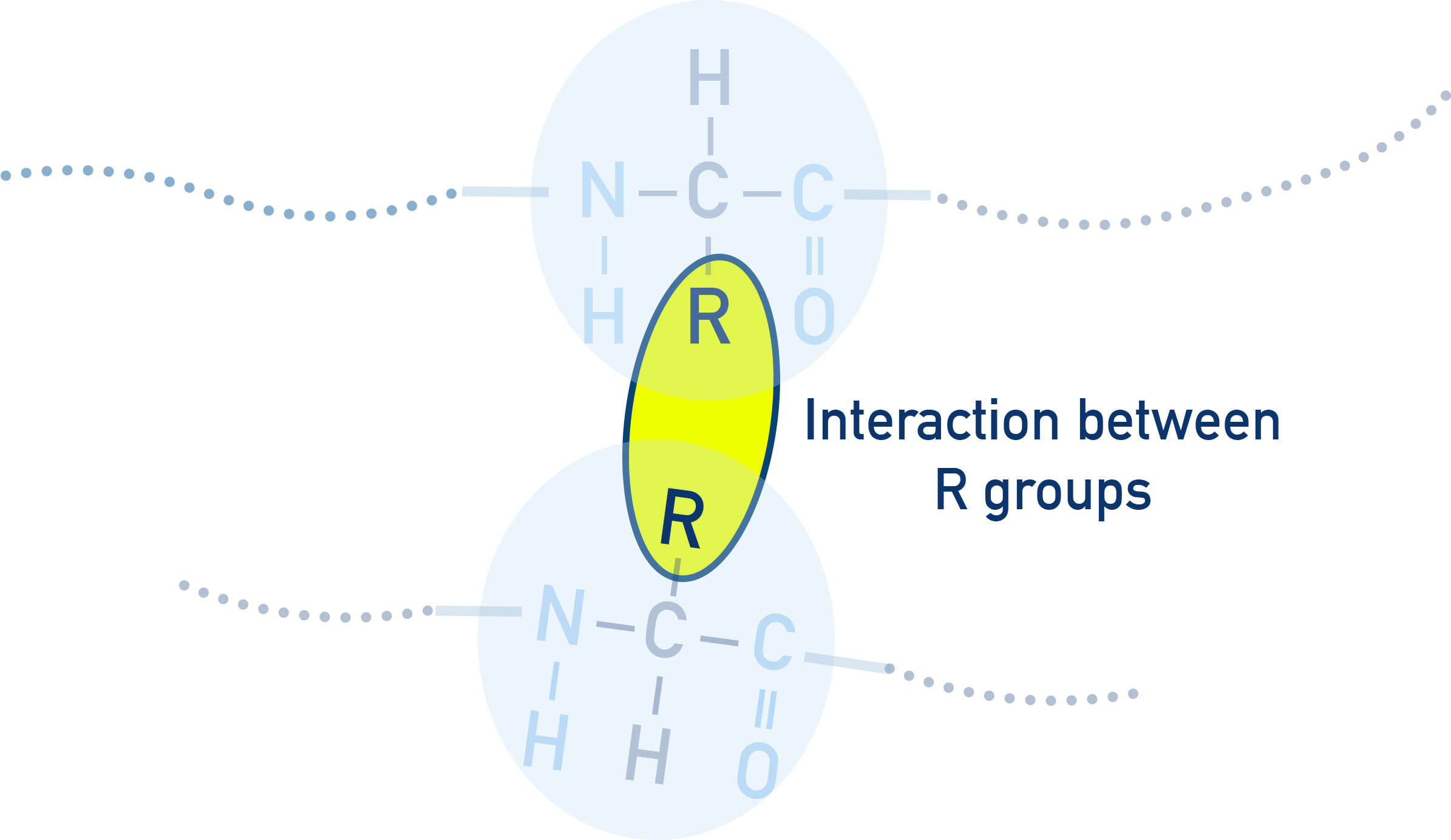
The interactions between different R groups causes the protein to ‘fold’ into complicated shapes, with the final shape between determined by the locations (and type) of amino acids in the chain.
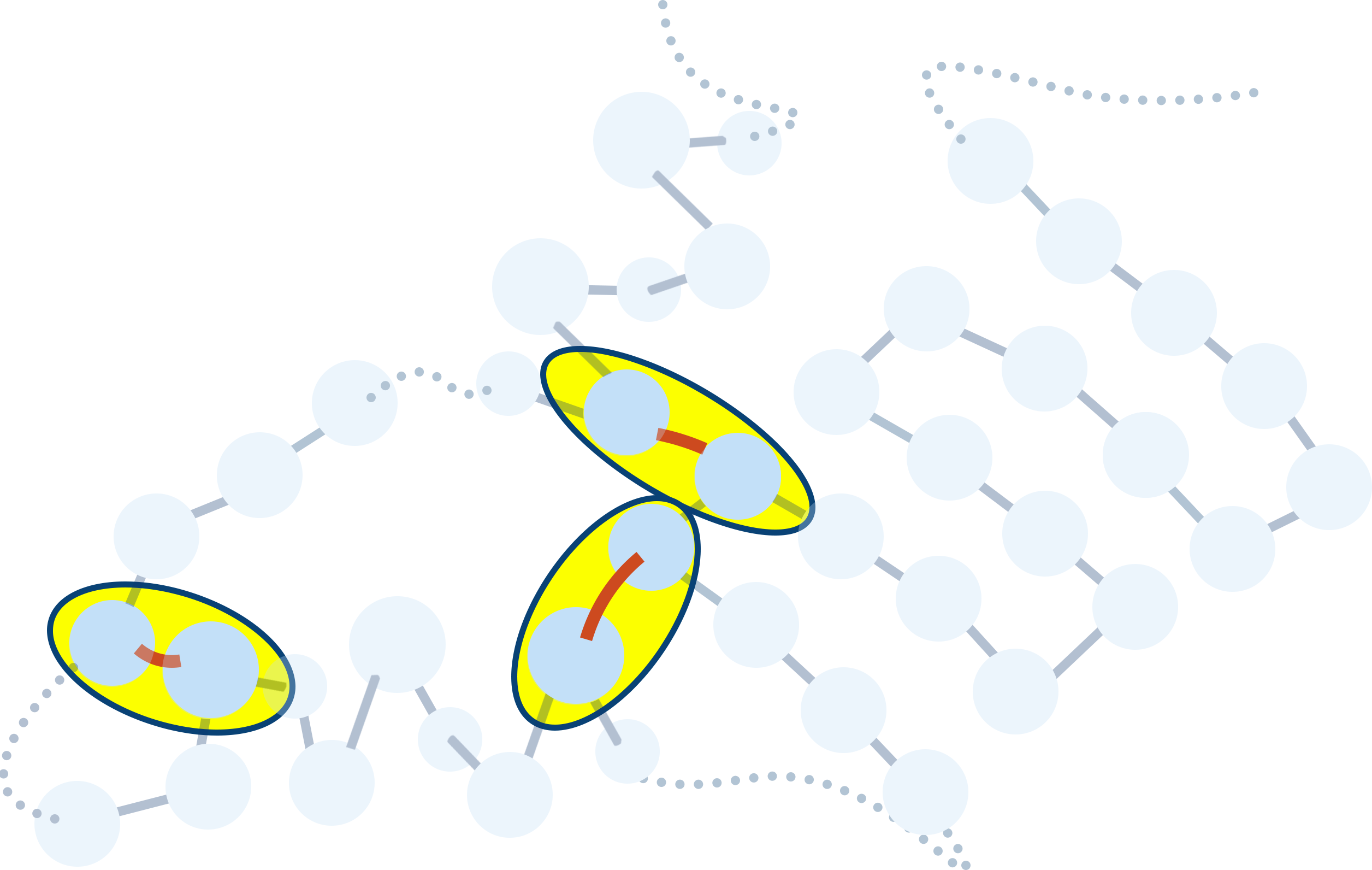
The type of interaction depends on the R groups interacting - only certain R groups can interact with each other.
- Hydrogen bonding (between polar groups that can form Hydrogen bonds).
- Disulfide (S−S) bonds (between cysteine residues that contain sulfur (S)).
- Ionic bonds (between charged R groups).
- Hydrophobic interactions (between non-polar R groups).
Hydrolysis of Proteins
Proteins are examples of condensation polymers and can be broken apart into amino acids by hydrolysis. See Condensation Polymers for more detail.
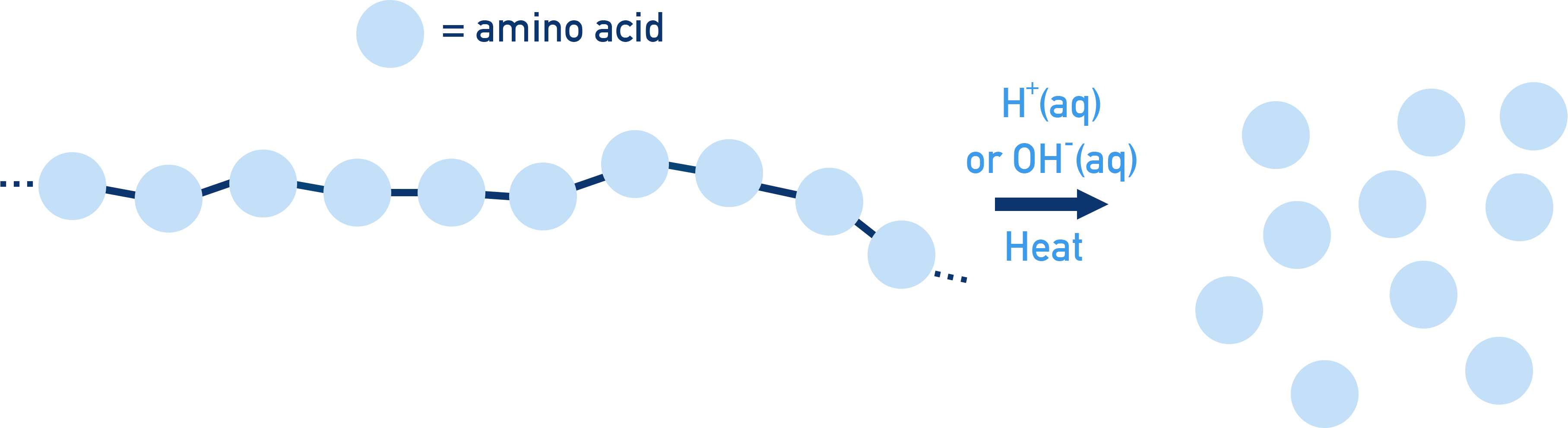
Peptide bonds (−CONH−) are hydrolysed by acid, alkali, or enzymes to release amino acids. The form of amino acid(s) produced is based on the conditions used (see Amino Acids for more detail).
Reagents:
- Acid hydrolysis: Heat with HCl under reflux.
- Alkaline hydrolysis: Heat with NaOH.
Separation and Identification of Amino Acids by Thin-Layer Chromatography (TLC)
After a protein is hydrolysed and broken apart into its amino acids, the amino acids can be separated and identified using Thin-Layer Chromatography (TLC) – see Required Practical 12.
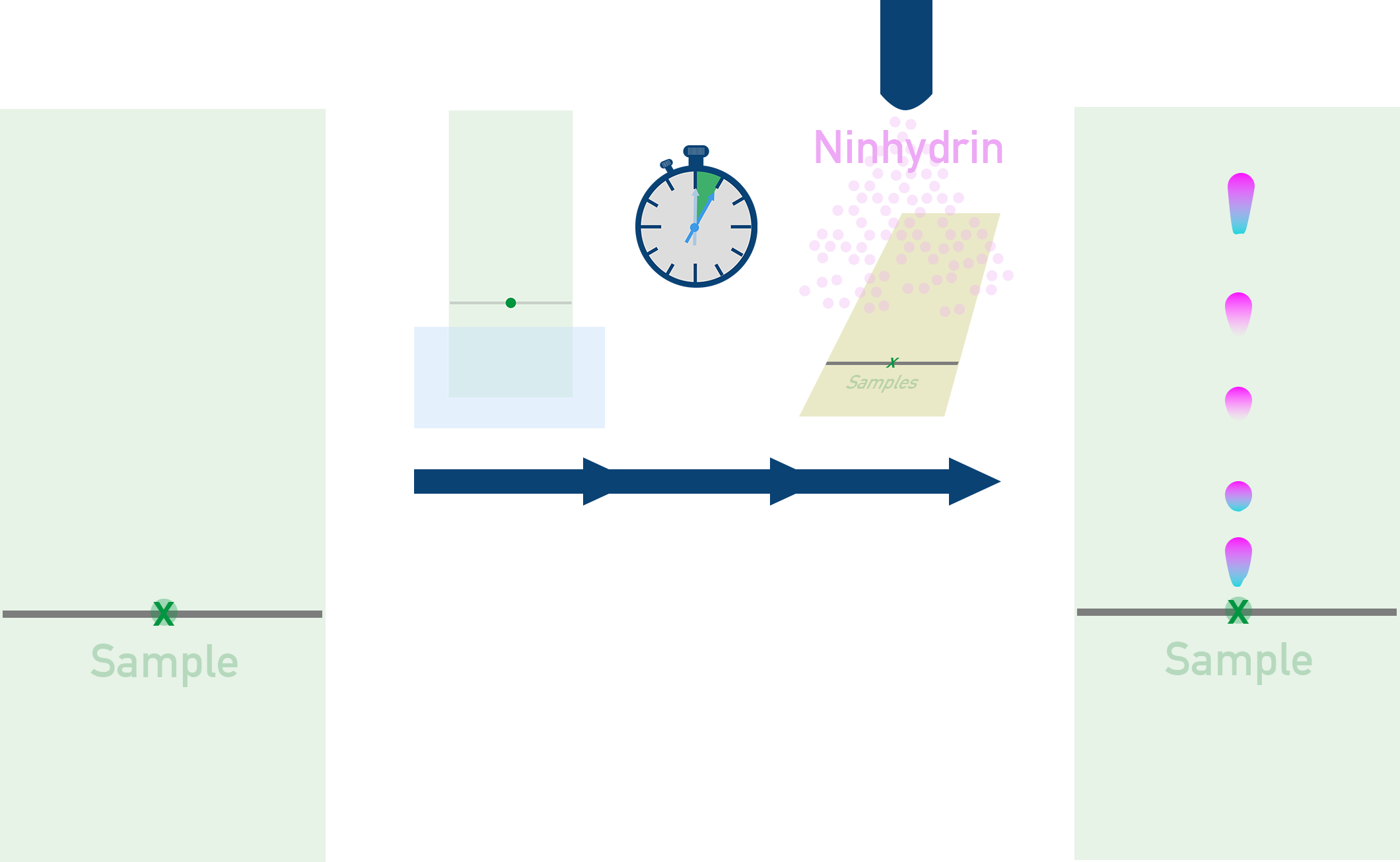
- Amino acids are separated based on their solubility in the solvent.
- Ninhydrin dye or UV light is used to locate amino acids on the chromatogram.
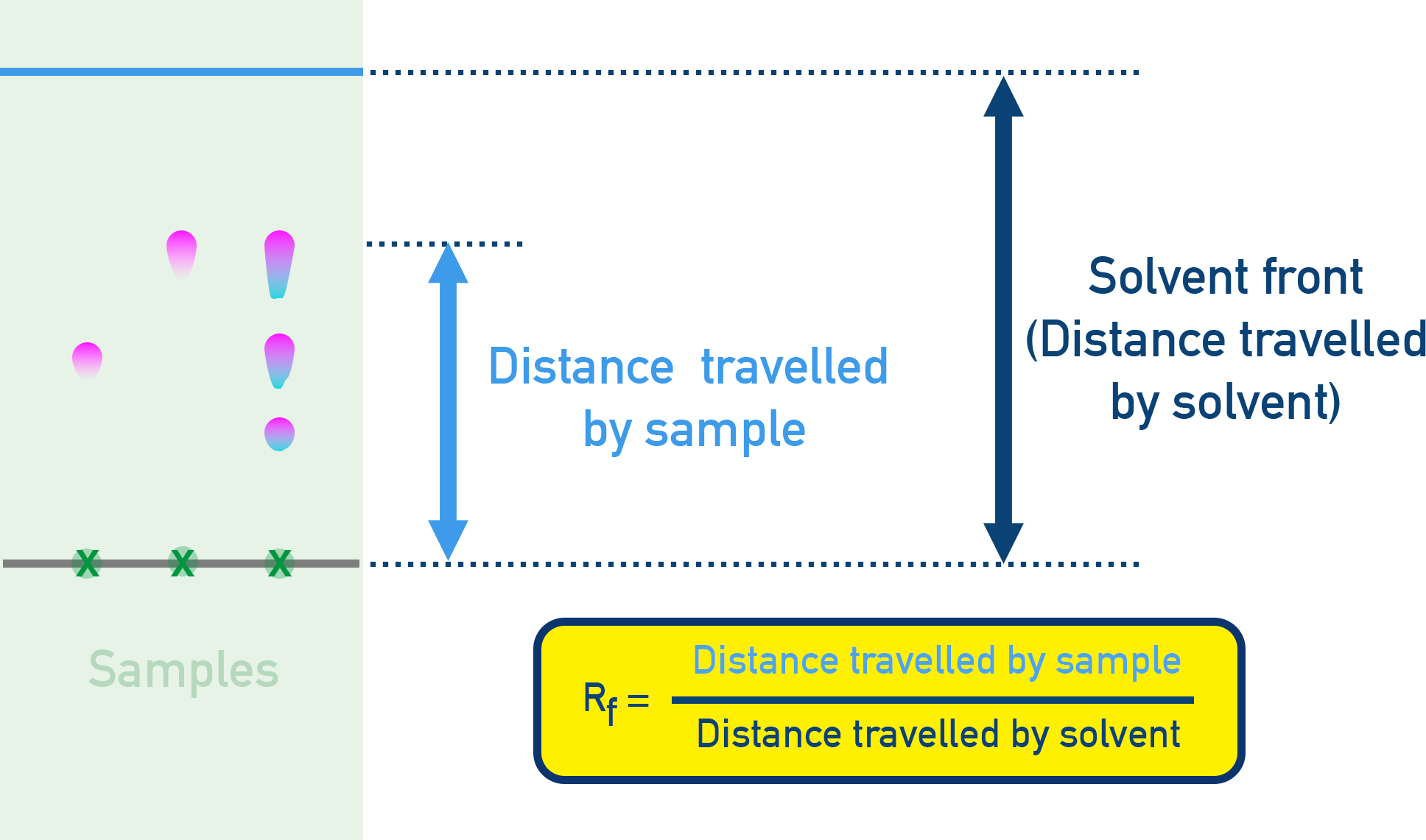
- Rf values (Retention Factor) are calculated: Rf = Distance moved by compound ÷ Distance moved by solvent.
- Each amino acid has a unique Rf value, allowing identification.
Summary
- Proteins are polymers of amino acids linked by peptide (−CONH−) bonds formed in condensation reactions.
- Primary structure = amino acid sequence; secondary = α-helix/β-sheet via hydrogen bonding; tertiary = overall 3D fold from R-group interactions.
- Protein hydrolysis (acid/alkali/enzymes) breaks peptide bonds to release amino acids.
- TLC separates amino acids; detect with ninhydrin/UV and identify using Rf values.
